Bridge Molecule and Life
Biological system is composed of molecules, cells, tissues, organs, etc with their hierarchical complicated interactions. Resulting from a stimulus on the microscopic level, the system can perform meso- and macroscopic functions robustly even under thermal-fluctuating environment. Such functions can be rationalized as a `sequence’ of structural changes involving chemical reactions triggered by the stimulus across hierarchies of time and space scales. There exist two distinct strategies to explore the mechanisms of such biological systems, that is, an anatomical bottom-up approach which builds the system from the microscopic molecular basis, and a constructive top-down approach in which one develops (phenomenological) models to capture some essential aspects of the biological systems. However, the former solely articulates the composite elements and the latter does not exclude possibilities which end up with models far apart from the reality because of the coarse-graining of the systems. The main purpose of our laboratory is to understand the fundamental principles of chance and necessity of “change of states”, and to construct new concepts and methodologies to bridge the gap between such top-down and bottom-up approaches for biological systems, enabling us to unveil the mechanisms that bridge molecules and life across hierarchies in time and space.
Research Topics
- Fundamental principle of selectivity and stochasticity in “changes of states” such as chemical reactions and conformational change of biomolecules
- Complexity of Protein Landscape, Conformation Network and Dynamics
- Developments of new methodologies and concepts to bridge molecules and life based on single molecule time series
- Information flow across hierarchies of time and space and its relation to biological functions
- Adaptability, robustness and emergence in complex systems
Fundamental principle of selectivity and stochasticity in “changes of states” such as chemical reactions and conformational change of biomolecules
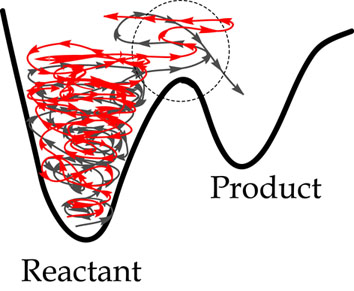
Chemical reactions are regarded as the most elementary prototype of change of the states or matter. How a rearrangement of atoms or molecules constituting a system occurs is one of the most intriguing fundamental questions from the day of alchemy. However why can the system react from reactants to the products? In other words, in the figure below, what origin differentiates the initial conditions which bring the system to the product (gray colored trajectory) or not (red colored trajectory)? So far, most of all chemical reaction theories divide the wholeness into the reacting system (part) and the residuals, which has been regarded as a heat bath and regard chemical reactions or conformation change of molecules as being governed by `casting dice’. However, recently we revealed using Lie canonical perturbation theory combined with microcanonical molecular dynamics simulation that normally hyperbolic invariant manifolds (NHIMs) and the stable and unstable invariant manifolds survive even in a sea of chaos, and this can provide a deterministic regularized reaction pathway to mediate chemical reaction in the `stochastic’ process of an n-particle system. Our findings indicate that nature may have, at least locally, known a priori the destination of chaotic reaction dynamics. The stability and bifurcation of the NHIM are thus the most essential subjects in understanding “who knows the goal of the reaction” and “what the whole and parts are in the change of the state.”
Books
- “Advancing Theory for Kinetics and Dynamics of Complex, Many-Dimensional Systems: Clusters and Proteins” Advances in Chemical Physics, 145 ed. by T. Komatsuzaki, R. S. Berry, D.M. Leitner John-Wiley & Sons, Inc. ISBN: 978-0-470-64371-6(2011)
- “Geometric Structures of Phase Space in Multi-Dimensional Chaos: Applications to Chemical Reaction Dynamics in Complex Systems” Advances in Chemical Physics, vol. 130A and 130B Ed. by M. Toda, T. Komatsuzaki, T. Konishi, R. S. Berry and S. A. Rice, John-Wiley & Sons, Inc. 1224 pages (2005) (References therein).
Selected Review Articles
- David M. Leitner, Yasuhiro Matsunaga, Akira Shojiguchi, Chun-Biu Li, Tamiki Komatsuzaki, and Mikito Toda “Non-Brownian Phase Space Dynamics of Molecules, the Nature of their Vibrational States, and non-RRKM Kinetics” Advances in Chemical Physics 145 in press.
- Shinnosuke Kawai, Hiroshi Teramoto, Chun-Biu Li, Tamiki Komatsuzaki, Mikito Toda, “Dynamical Reaction Theory based on Geometric Structures of Phase Space” Advances in Chemical Physics 145 in press.
- Tamiki Komatsuzaki and R. Stephen Berry,’Chemical Reaction Dynamics: Many-Body Chaos and Regularity’ Advances in Chemical Physics123,79-152, Ed. by I. Prigogine and S. A. Rice, John-Wiley & Sons, Inc. (2002)
- Komatsuzaki, T. & Berry, R. S. (2005) Regularity in Chaotic Transitions on Two-Basin Landscapes. Advances in Chemical Physics130A, 143-170. (Toda, M., Komatsuzaki, T., Konishi, T., Berry, R.S. & Rice, S.A. eds.) John Wiley & Sons, Inc. New York.
- a short review article rejected by a Journal after 4 months’ reviewing (1999)
- the corresponding Appendix requested by the Journal(1999)
- a short review article rejected by a Journal after 7 months’ reviewing (2009.1.31-2009.9.10)[its supporting infomation]
Selected Representative Publications
- Hiroshi Teramoto, Mikito Toda, Tamiki Komatsuzaki, “A Dynamical Switching of a Reaction Coordinate to Carry the System Through to a Different Product State at High Energies” Physical Review Letters 106, 054101 (2011) [4 pages].
- Shinnosuke Kawai and Tamiki Komatsuzaki ‘Robust existence of a reaction boundary to separate the fate of a chemical reaction’ Physical Review Letters 105, 048304 (2010).
- Shinnosuke Kawai and Tamiki Komatsuzaki, “Phase Space Geometry of Dynamics Passing through Saddle Coupled with Spatial Rotation” Journal of Chemical Physics 134, 084304 (2011)[11 pages].
- Shinnosuke Kawai and Tamiki Komatsuzaki ‘Quantum Reaction Boundary to Mediate Reactions in Laser Fields’ Journal of Chemical Physics 134, 024317 [13 pages] (2011).
- Shinnosuke Kawai and Tamiki Komatsuzaki : `Dynamic pathways to mediate reactions buried in thermal fluctuations. I. Time-dependent normal form theory for multidimensional Langevin equation’, Journal of Chemical Physics, 131 : 224505-11pages (2009).
- Chun Biu Li, Mikito Toda, and Tamiki Komatsuzaki ‘Bifurcation of no-return transition states in many-body chemical reactions’ Journal of Chemical Physics 130, 124116 (2009).
- Hiroshi Teramoto and Tamiki Komatsuzaki ‘Exploring Remnant of Invariants Buried in a Deep Potential Well in Chemical Reactions’ Journal of Chemical Physics 129, 094302 (2008)
- Akira Shojiguchi, Chun Biu Li, Tamiki Komatsuzaki, and Mikito Toda ‘Fractional behavior in nonergodic reaction processes of isomerization’ Physical Review E (rapid communication) 75, 035204(R) (2007).
- Shinnosuke Kawai, Yo Fujimura, Okitsugu Kajimoto, Takefumi Yamashita, Chun-Biu Li, Tamiki Komatsuzaki, and Mikito Toda ‘Dimension reduction for extracting geometrical structure of multidimensional phase space: Application to fast energy exchange in the reaction O(1D)+N2O →NO+NO ‘ Physical Review A 75, 022714 (2007) (11 pages).
- Chun Biu Li, Akira Shojiguchi, Mikito Toda, and Tamiki Komatsuzaki ‘Definability of No-return Transition States in High Energy Regime Above Threshold’ Physical Review Letters 97, 028302 (2006).
- Chun Biu Li, Yasuhiro Matsunaga, Mikito Toda, and Tamiki Komatsuzaki ‘Phase Space Reaction Network on a Multisaddle Energy Landscape: HCN isomerization’ Journal of Chemical Physics 123, 184301(13)(2005).
- Tamiki Komatsuzaki and R. Stephen Berry, ‘A Dynamical Propensity Rule of Transitions in Chemical Reactions’ Journal of Physical Chemistry A 106,10945-10950(2002)
- Tamiki Komatsuzaki and R. Stephen Berry ‘Dynamical Hierarchy in Transition States: Why and How Does a System Climb over the Mountain?’ Proceedings of National Academy of Sciences USA,(2001) 98,7666-7671
- Tamiki Komatsuzaki and R. Stephen Berry ‘Regularity in Chaotic Reaction Path I: Ar6’ Journal of Chemical Physics (1999), 110(18), 9160-9173
- Tamiki Komatsuzaki and R. Stephen Berry ‘Regularity in Chaotic Reaction Path II: Ar6 -Energy Dependence and Visualization of the Reaction Bottleneck-‘ Physical Chemistry Chemical Physics (1999) 1, 1387-1397
- Tamiki Komatsuzaki and Masataka Nagaoka ‘Study on “Regularity” of the Barrier Recrossing Motion’, Journal of Chemical Physics(1996) 105,10838-10848
Complexity of Protein Landscape, Conformation Network and Dynamics
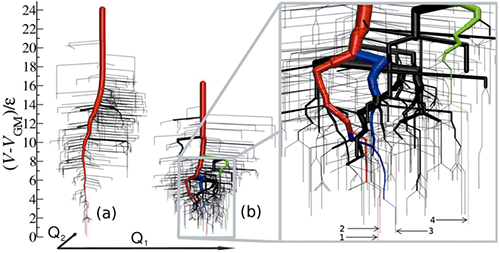
Why can natural proteins find their unique native state among the candidates of states, whose number is estimated to be beyond the age of the universe? The key concept of protein folding problem has been interpreted as the `design’ of the protein landscapes that the protein might have acquired via mutations. Natural (small) proteins are believed to be designed as having a funnel-like energy landscape introducing a strong energy bias toward the global minimum. However, how can one quantify the degree of ruggedness or multiplicity of paths in terms of the underlying multi-dimensional energy landscape? What is the environment for protein in solutions, cells, or organisms? Can we put all the effect of the environment, e.g., water molecules in the vicinity of protein molecules, into the thermal bath and the friction source? We have been developing a new platform to resolve the important contemporary issues in complexity of protein landscape and dynamics in terms of several advanced techniques in data mining (principal component analysis), information theory, network theory and fluid mechanics (water vector field analysis, Lagrangian coherent structure).
Books
- “Advancing Theory for Kinetics and Dynamics of Complex, Many-Dimensional Systems: Clusters and Proteins” Advances in Chemical Physics, 145 ed. by T. Komatsuzaki, R. S. Berry, D.M. Leitner John-Wiley & Sons, Inc. ISBN: 978-0-470-64371-6(2011)
Selected Review article
- Tamiki Komatsuzaki, Akinori Baba, Shinnosuke Kawai, Mikito Toda, John E. Straub, R. Stephen Berry “Ergodic Problems for Real Complex Systems in Chemical Physics” Advances in Chemical Physics 145, in press.
- Akinori Baba and Tamiki Komatsuzaki ‘Multidimensional Energy Landscapes in Single Molecule Biophysics’ Advances in Chemical Physics 146, in press.
- C. B. Li and T. Komatsuzaki : “Extracting the Underlying Unique Reaction Scheme from a Single-Molecule Time Series” in Cell Signaling Reactions: Single-Molecular Kinetic Analysis (Springer) ISBN: 978-90-481-9863-4.(2011) (invited)
- Tamiki Komatsuzaki, Kyoko Hoshino and Y. Matsunaga, ‘Regularity in Chaotic Transitions on Multi-Basin Landscapes ‘ Advances in Chemical Physics 130B, pp257-313 (2005) eds. by M. Toda, T. Komatsuzaki, T. Konishi, R.S. Berry and S. A. Rice, John-Wiley & Sons, Inc.
Selected Relevant publications
- Akinori Baba and Tamiki Komatsuzaki ‘Extracting the Underlying Effective Free Energy Landscape From Single-Molecule Time Series — Local Equilibrium States and their Network’ Physical Chemistry Chemical Physics, 13 (4), 1395 – 1406 (2011).
- Akinori Baba and Tamiki Komatsuzaki ‘Construction of effective free energy landscape from single molecule time series’ Proceedings of National Academy of Sciences USA 104(49),19297-19302 (2007).
- Yasuhiro Matsunaga, Chun Biu Li, and Tamiki Komatsuzaki ‘Anomalous Diffusion in Folding Dynamics on Minimalist Protein Landscape’ Physical Review Letters 99, 238103 (2007) .
- Yasuhiro Matsunaga, Konstantin S. Kostov and Tamiki Komatsuzaki, ‘Multi-Basin Dynamics in Off-Lattice Minimalist Protein Landscapes’ Journal of Physical Chemistry A 106,10898-10907 (2002)
- Tamiki Komatsuzaki, Kyoko Hoshino, Yasuhiro Matsunaga, Gareth J. Rylance, Roy L. Johnston and David J. Wales ‘How Many Dimension is Required to Approximate Potential Energy Landscape of A Model Protein?’ Journal of Chemical Physics 122, 084714-1~084714-9 (2005).
- Gareth J. Rylance, Roy L. Johnston, Yasuhiro Matsunaga, Chun Biu Li, Akinori Baba,Tamiki Komatsuzaki, Topographical complexity of multidimensional energy landscapes, Proceedings of National Academy of Sciences USA 103, 18551 (2006)
Developments of new methodologies and concepts to bridge molecules and life based on single molecule time series
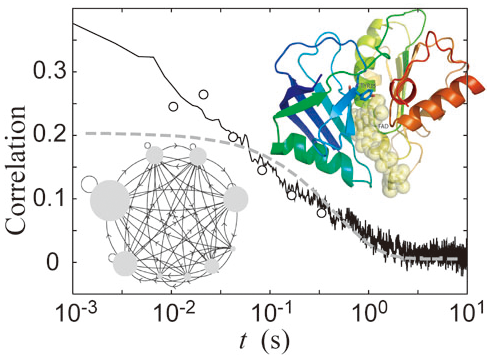
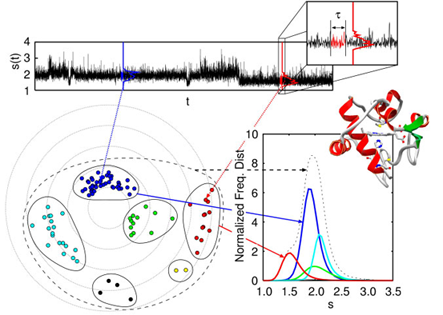
Optical single-molecule spectroscopy has provided a new scope in biology with unique insights into both the distribution of molecular properties and their dynamic behavior at single molecule level, which are inaccessible by ensemble-averaged measurements. These techniques enable us to explore the mechanism of biological functions under the competition with thermal fluctuation in single molecule level. If people could access the detailed information of molecular dynamics in terms of computer simulation at the day of Boltzmann and Gibbs, people might have devoted most of their time in order to establish the ergodicity hypothesis in the stream of the actual multivariate data, and might not have come up with the idea of constructing statistical mechanics. In this sense, the observation of single molecule events should provide a big challenge in exploring the theories of functions of biological systems. Our main aim here is to explore what one can learn from the single-molecule time series about the underlying states and their network of the system and/or free energy landscapes, whose topology and topography should depend on the time scale of observation. We want to establish the novel dynamical techniques and concepts to bridge molecules and life in terms of single molecule time series and provide a new perspective for addressing the nature of their hierarchical organization in multidimensional state space.
Books
- “Advancing Theory for Kinetics and Dynamics of Complex, Many-Dimensional Systems: Clusters and Proteins” Advances in Chemical Physics, 145 ed. by T. Komatsuzaki, R. S. Berry, D.M. Leitner John-Wiley & Sons, Inc. ISBN: 978-0-470-64371-6(2011)
- “Single Molecule Biophysics: Experiments and Theories” Advances in Chemical Physics, 146 ed. by T. Komatsuzaki, S. Takahashi, M. Kawakami, Haw Yang and Robert Silbey, John-Wiley & Sons, Inc. ISBN: 978-1-1180-5780-3(2011).
Selected Review article
- C. B. Li and T. Komatsuzaki : “Extracting the Underlying Unique Reaction Scheme from a Single-Molecule Time Series” in Cell Signaling Reactions: Single-Molecular Kinetic Analysis (Springer) ISBN: 978-90-481-9863-4.(invited) .
- Akinori Baba and Tamiki Komatsuzaki ‘Multidimensional Energy Landscapes in Single Molecule Biophysics’ Advances in Chemical Physics 146, in press.
- Tamiki Komatsuzaki, Akinori Baba, Shinnosuke Kawai, Mikito Toda, John E. Straub, R. Stephen Berry “Ergodic Problems for Real Complex Systems in Chemical Physics” Advances in Chemical Physics 145, in press.
Selected Relevant publications
- Akinori Baba and Tamiki Komatsuzaki ‘Extracting the Underlying Effective Free Energy Landscape From Single-Molecule Time Series — Local Equilibrium States and their Network’ Physical Chemistry Chemical Physics, 13 (4), 1395 – 1406 (2011).
- Shinnosuke Kawai and Tamiki Komatsuzaki, “Derivation of the Generalized Langevin Equation in Non-stationary environments” Journal of Chemical Physics 134, 114523 (2011)[12 pages].
- Chun Biu Li, Haw Yang, and Tamiki Komatsuzaki: “New Quantification of Local Transition Heterogeneity of Multiscale Complex Networks Constructed from Single-Molecule Time Series”, Journal of Physical Chemistry B, 113 (44), 14732–14741 (2009)
- Chun Biu Li, Haw Yang, and Tamiki Komatsuzaki ‘Multiscale Complex Network of Protein Conformational Fluctuation Buried in Single Molecule Time Series’ Proceedings of National Academy of Sciences USA,105, 536-541 (2008).
- Akinori Baba and Tamiki Komatsuzaki ‘Construction of effective free energy landscape from single molecule time series’ Proceedings of National Academy of Sciences USA 104(49),19297-19302 (2007).
- Masahito Kinoshita, Kiyoto Kamagata, Akio Maeda, Yuji Goto, Tamiki Komatsuzaki and Satoshi Takahashi ‘Development of a technique for the investigation of folding dynamics of single proteins for extended time periods’ Proceedings of National Academy of Sciences USA 104, 10453(2007)
- Shinnosuke Kawai and Tamiki Komatsuzaki : `Dynamic pathways to mediate reactions buried in thermal fluctuations. I. Time-dependent normal form theory for multidimensional Langevin equation’, Journal of Chemical Physics, 131: 224505-11pages (2009).
Information flow across hierarchies of time and space and its relation to biological functions
Biological functions such as signal transduction from the periplasmic to cytoplasmic space result from a sequence of structural changes involving chemical reactions triggered by a stimuli. The biological functions should inherently accompany information flow across hierarchies of time and space scales under thermally-fluctuating environments. The existence of information flow across different time and space scales implies the breakdown of the concept of adiabaticity, in the other words, the assumption of the existence of separation of time scale. For instance, the concept of free energy landscape often used in protein science implicitly assumes the existence of the separation of time scale between the degrees of freedom that describe the landscape and the other degrees of freedom responsible for what people call “thermal bath”. One of the unaware important aspects is, for example, that the composition of degrees of freedom that belong either to describing the landscape and to the thermal bath, and the morphology and dimensionality of the landscape should change through the course of the signal transduction taking place across different time scale. Yet, it is far from the situation that one could compute such events in the firm molecular basis. Our aim is to construct a technique to quantify the causal relation or the information flow in terms of multiscale complex network or multiscale free energy landscape of variable dimension constructed on the basis of single-molecule time series, and reveal the signal transduction pathway in terms of the underlying state space or energy landscape.
Adaptability, robustness and emergence in complex systems
Why and how can biological systems adapt and sometimes even “reprogram” the functions of genes or cells when they are under the nonperturbative change of environment? The adaptability, robustness, and emergence against the changes of the environment that most of all biological systems can naturally resolve are far from the understanding of their physical foundation. The adaptability of the systems may be interpreted as the changes of the underlying state space network or multiscale free energy landscapes along the course of the events. For instance, the differences of the dimensionality and topographical features of the state space network constructed from single molecule time series before and after the changes are expected to shed light on the origin of adaptability. The robustness of functions in multiscale biological systems is subject to how the information can persist across the different time and space scales under the thermally-fluctuating environment beyond the concept of the so-called slaving principle in synergetics. Our purpose is to establish the techniques that enable us to explore the origin of adaptability, robustness, and emergence in terms of molecular basis using single molecule time series of biological systems, and to provide several clues in the construction of the corresponding mathematical framework that bridges molecule and life.
